Nucleic acids (DNA and RNA), DNA Extraction, Cell lysis, Color test for ribose and deoxyribose
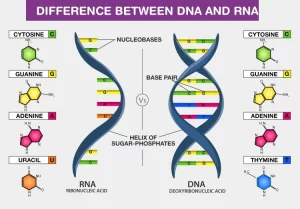
DNA is the site of storage of genetic material. It is mainly present in the nucleus (nuclear DNA). In addition, mitochondrial DNA also exists, carrying a much less number of genes, mainly concerned with the respiratory chain. RNA is the nucleic acid responsible for converting genes to proteins.
Nucleic acids (DNA and RNA)
DNA is highly compacted inside the nucleus, and is wrapped around histone proteins. The negatively charged DNA finds it easy to adhere to the positively charged histones, DNA is enclosed within the nuclear membrane and the two DNA strands are interconnected to each other by hydrogen bonds.
Color tests for nucleic acid components
Complete hydrolysis of a nucleic acid gives a mixture of 4 constituent bases, a pentose sugar, and one mole of phosphoric acid per mole of the base. Various chemical conditions as well as specific enzymes have been employed for partial hydrolysis to particular fragments, such as nucleosides, nucleotides, ribose phosphates, and free bases. The color tests on RNA and DNA given below are carried out under hot, acidic conditions, which will affect partial or complete hydrolysis of the nucleic acids.
Color test for ribose
A. Mix 0.5ml of RNA with 2ml orcinol reagent in the test tube. Boil the tube in a boiling water bath for 10-15 min, a green color is formed.
Color test for deoxyribose
B. Mix 1 ml of DNA with 2 ml of diphenylamine reagent. Boil for 10 min in a boiling water bath, a blue color is formed.
DNA Extraction
To extract DNA from within the cell, Some common steps have to be done.
Cell lysis
1. Breaking the cells, commonly referred to as cell disruption cell lysis, to expose the DNA within. This is commonly achieved by chemical and physical methods such as blending, grinding or sonicating the sample.
The lysis solution contains two main ingredients:
- Detergent or surfactant which dissolves the cell membrane and nuclear envelope causing the cell to burst open and release the DNA.
- Proteinase k for removing proteins.
2. Removing RNA by adding an RNase (almost always done)…
3. Add concentrated salt solution such as ammonium or sodium acetate, the salts cause Precipitation of the protein and clumping of other cellular debris together.
4. Centrifugation at high speed, Heavy clumps of proteins and cellular debris sink to the bottom of the tube, while the strands of DNA remain distributed through the liquid.
5. Carefully remove the top liquid (which contains the DNA) into a clean tube.
6. DNA purification from detergents, proteins, salts, and reagents used during cell lysis step:
- Ethanol precipitation is usually by ice-cold ethanol or isopropanol, Since DNA is insoluble in these alcohols, it will aggregate together, giving a pellet upon centrifugation, Precipitation of DNA is improved by increasing of ionic strength, usually by adding sodium acetate.
- Phenol-chloroform extraction in which phenol denatures proteins in the sample, In brief, aqueous samples are mixed with an equal volume of phenol: chloroform mixture. After centrifugation of the sample, two distinct phases are formed due to phenol and chloroform being miscible but having different densities, The denaturated proteins will partition into the lower organic phase while The DNA will be found at the interface between the two phases, DNA is then precipitated by mixing with cold ethanol or isopropanol and then centrifuging, For DNA isolation, phenol was used buffered to pH 8, RNA must be isolated using acidic phenol).
- Minicolumn purification that relies on the fact that the nucleic acid may bind (adsorption) to the solid phase (silica or other) depending on the pH and the salt content of the buffer.
7) After isolation, the DNA is dissolved in a slightly alkaline buffer, usually in the TE (Tris- EDTA) buffer, or in ultra-pure water and stored at 4 °C for further manipulation or at – 20 °C for long-term storage, The experiment in this practical section demonstrates a rather simple, yet effective, technique to extract DNA from tomatoes.
Extraction procedure
Items needed for the experiment:
- Tomatoes.
- Petri dish, plastic knife, funnel, gauze, test tube, dropper.
- Soap detergent.
- Table salt.
- EDTA.
- Cold alcohol
Steps
1- In a petri dish, peal the tomatoes and crush them using a plastic knife, this step aims to break open (LYSE) the cell wall, cell membrane, and the nuclear envelope to allow the DNA to be released.
2- Add a soap detergent to the crushed tomatoes. Leave for 10 minutes, this step aims to break down the fat and proteins that make up the cell and nuclear membrane. (Just like the detergent dissolves grease on your dishes).
3- Table salt and EDTA salt are added to the mixture, DNA has a negative electrical charge due to the phosphate groups on the DNA backbone, and the electrical charge makes it soluble, When salt is added to the sample, the positively charged sodium ions of the salt are attracted to the negative charges of the DNA, neutralizing the electrical charge of the DNA, This allows the DNA molecules to come together instead of repelling each other, EDTA chelates Mg to inhibit nuclease enzymes and stabilize DNA.
4- Transfer the mixture to a clean test tube using a funnel and gauze. Add ice-cold alcohol drop by drop using a dropper to the mixture and observe. DNA does not dissolve in alcohol. (It is soluble in water), The addition of alcohol causes the DNA to precipitate, Ethanol is less dense than water so it floats on top. All of the proteins we broke up will sink to the bottom; while the DNA will float on top.
Extracted DNA will appear like cotton clouds in the test tube and can be picked up using a toothpick, Extracted DNA can be used in several applications such as PCR, electrophoresis, restriction fragment length polymorphism…ect as will be discussed.
Nucleic acid quantitation
In molecular biology, quantitation of nucleic acids is commonly performed to determine the average concentrations of DNA or RNA present in a mixture, as well as their purity, Reactions that use nucleic acids often require particular amounts and purity for optimum performance, There are several methods to establish the concentration of a solution of nucleic acids, including spectrophotometric quantification and UV fluorescence in the presence of a DNA dye.
Detection of nucleic acid purity using 260/280 ratio
The ratio of absorbance at 260 nm and 280 nm is used to assess the purity of DNA and RNA, A ratio of ∼ 1.8 is generally accepted as “pure” for DNA; a ratio of ~2.0 is generally accepted as “pure” for RNA, If the ratio is appreciably lower in either case, it may indicate the presence of protein, phenol or other contaminants that absorb strongly at or near 280 nm.
Quantitation of nucleic acids using a spectrophotometer
Use H₂O or IX TE as a solvent to suspend the nucleic acids, and place each sample in a quartz cuvette, Zero the spectrophotometer with a sample of solvent, For more accurate readings of the nucleic acid sample of interest, dilute the sample to give readings between 0.1 and 1.0. For a 1-cm pathlength, the optical density at 260 nm (OD260) equals 1.0 for the following solutions:
- 50 µg/mL solution of dsDNA.
- 33 µg/mL solution of ssDNA.
- 20-30 µg/mL solution of oligonucleotide.
- 40 μg/mL solution of RNA.
Contamination of nucleic acid solutions makes spectrophotometric quantitation inaccurate, Calculate the OD260/OD280 ratio for an indication of nucleic acid purity.
You can subscribe to Science Online on YouTube from this link: Science Online
You can download the Science Online application on Google Play from this link: Science Online Apps on Google Play
Cloning of DNA sequences, Importance of recombinant DNA & Uses of human genome
Molecular technology (Genetic Engineering) advantages, Techniques and uses
Importance of Nucleosides, Nucleotides, Purines, Pyrimidines & Sugars of nucleic acids
Regulation of the cell cycle, DNA synthesis phase, Interphase & Mitosis