Control of Drug Resistance and Bacterial resistance to chemotherapeutic agents
Drug resistance is the reduction in the effectiveness of the medication such as the antimicrobial or the antineoplastic in treating the disease or condition, Drug resistance is used in the context of resistance that pathogens or cancers have “acquired”, that is, resistance has evolved, Antimicrobial resistance and antineoplastic resistance challenge clinical care and drive research, If the organism is resistant to more than one drug, it is said to be multidrug-resistant.
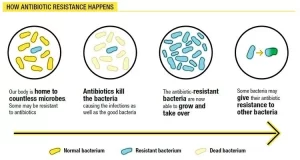
Bacterial resistance to chemotherapeutic agents
There are many different mechanisms by which microorganisms might exhibit resistance to drugs:
1. Microorganisms produce enzymes that destroy the active drug, e.g., staphylococci resistant to penicillin G produce a β-lactamase that destroys the drug, Bacteria resistant to aminoglycosides produce adenylating, phosphorylating or acetylating enzymes that destroy the drug, Bacteria may be resistant to chloramphenicol if they produce a chloramphenicol acetyltransferase.
2. Microorganisms reduce their permeability to the drug. In resistant cells, the drug is either not actively transported into the cell or leaves it so rapidly (by efflux pump) that inhibitory concentrations are not maintained. Examples include tetracyclines, aminoglycosides, and polymyxins.
3. Microorganisms develop an altered structural target for the drug, e.g., chromosomal resistance to aminoglycosides is associated with the loss or alteration of a specific protein on the 30 S subunit of the bacterial ribosome, Erythromycin-resistant organisms have an altered receptor on the 50S subunit of the ribosome Resistance to some penicillins may be a function of the loss or alteration of PBPS.
4. Microorganisms develop an altered metabolic pathway that bypasses the reaction inhibited by the drug, e.g., some sulphonamide-resistant bacteria do not require extracellular PABA but, like mammalian cells, can utilize preformed folic acid.
Origin of Drug Resistance
1. Non-genetic origin (natural or inherent): This is an innate property of the bacterium, so it is predictable. It is often due to the absence of a target structure for drug action (e.g absence of a cell wall in Mycoplasma). It may also be due to the presence of a permeability barrier in the cell envelope (e.g. Pseudomonas aeruginosa has a complex cell wall that limits penetration of many antibacterial agents). Also, microorganisms that are metabolically inactive (non-multiplying) may be resistant to drugs e.g. mycobacteria often survive in tissues for many years after infection and do not multiply.
2. Genetic origin (acquired): by methods that allow gene change or exchange. An organism may lose its susceptibility to an antibiotic during the course of treatment. so it is unpredictable. This may be due to chromosomal resistance, and extrachromosomal resistance.
Chromosomal resistance: which develops as a result of spontaneous mutation in a locus that controls susceptibility to a given antimicrobial drug. Spontaneous mutation occurs with a frequency of 10-8 to 10-12 and thus is an infrequent cause of the emergence of clinical drug resistance in a given patient. Chromosomal mutations are most commonly resistant by virtue of a change in a structural receptor for a drug or an alteration of a metabolic pathway.
Extrachromosomal resistance: Carried on plasmid or transposon genes. These genes often control the formation of enzymes capable of destroying antimicrobial drugs. Thus, plasmids that determine resistance to penicillins and cephalosporins carry genes for the production of ß-lactamases. Plasmids that carry resistance to chloramphenicol code for enzymes that destroy it (acetyltransferase).
Plasmids can also code for enzymes that acetylate, adenylate, or phosphorylate various aminoglycosides Other resistance genes that may be carried on extrachromosomal elements include genes that code for enzymes that determine the active transport of antibiotics e.g. tetracyclines across the cell membrane (reducing the import and or increasing the export of the drug).
High-level and Low-level resistance
High-level resistance refers to the resistance that cannot be overcome by increasing the dose of antibiotic. It is usually due to the enzyme-mediated destruction of the drug. A different antibiotic usually from another class should be used. Low-level resistance refers to the resistance that can be overcome by increasing the dose of antibiotic. It is usually due to mutations in the genes encoding a drug target, as the altered target can still bind some of the drugs but with reduced strength.
Cross Resistance: Microorganisms resistant to a certain drug may also be resistant to other drugs that share a mechanism of action. Such a relationship exists mainly between agents that are closely related chemically (e.g. polymyxin B and polymyxin E (colistin]. erythromycin and oleandomycin neomycin and kanamycin).
Control of Drug Resistance
The emergence of drug resistance in infections may be minimized in the following ways:
- Antibacterial drugs should not be prescribed without valid indication. They should be selected according to the proven or anticipated susceptibility of the infecting organism.
- Maintain high levels of the drug in the tissue for a long time to inhibit both the existing bacterial population and the first-generation mutants.
- Simultaneously administer two drugs, each of which delays the emergence of mutants. resistant to the other drug
- Avoid exposure of microorganisms to a particularly valuable drug by restricting its use.
You can follow science online on YouTube from this link: Science online
You can download Science online application on Google Play from this link: Science online Apps on Google Play
Antibiotics advantages, disadvantages, resistance and uses
Features and classification of viruses, Defective viruses and Viral vectors used for gene therapy
Staphylococci definition, diagnosis of staphylococcal infections & treatment
Antibacterial agents types, Effect of Antibacterial Drugs & How do antibiotics work?
Microbiology, Bacteria structure, types, Gram-positive bacteria & Gram-negative bacteria